Practical considerations for genomic epidemiology in public health
Roles, responsibilities, and most importantly, relationships
Genomic epidemiology: use of pathogen genomic data to determine the distribution and spread of an infectious disease in a specified population and the application of this information to control health problems
Creating a plan for building genomic epidemiology capacity in a public health agency is a key step in setting up a program and agency for success. Genomic epidemiology is highly interdisciplinary and overlaps with both the domains of bioinformatics and applied epidemiology. Without creating consensus between these groups on roles and responsibilities, it is easy to introduce friction as new staff are added and may be perceived as duplicative of existing staff’s spheres of responsibility.
In this article, I’ll address a few consideration when first building and structuring a genomic epidemiology program in a public health agency. My experience is primarily at a US state level, so while these principles may be helpful at all levels, that is the background I’m using to inform my Saturday morning musings on the topic.
Key decision points
Program location - where will genomic epidemiologists sit on the organizational chart
There are three primary options for where a genomic epidemiologists sits organizationally:
With the laboratory / bioinformatics group
With the epidemiologists
Multiple positions with some on each side (lab + epi)
Which answer is best will look different for every agency. Some considerations are:
Data access: A genomic epidemiologist will be unable to fulfill their role if they do not have access to both the sequencing data, and epidemiological data systems. If there are institutional barriers that preclude someone in the laboratory from accessing epi data systems, those either need to be addressed, or the genomic epidemiologist should be housed on the epidemiology side.
Human resources: As government agencies there can be policy, HR, or union-related reasons why certain job classses cannot report to others, so make sure there are no barriers for example with someone classified as an epidemiologist reporting to a laboratory scientist. There are ways to get around this (I often had one title on paper and in the state HR system, but another ‘working title’ that described what I actually did), but it’s good to explore.
Existing capabilities: Where will the new program or positions have the most support? If you have a strong existing bioinformatics program and they are eager to expand more into data integration and educating the applied epidemiologists, then this may be a good fit for your genomic epidemiologist. If you have an existing epidemiology lead with extensive background in molecular biology or bioinformatics, the program may be a good fit on the epi side.
Program goals: These will change over time as your program is staffed, but it’s good to have a framework as you go into hiring. If your first challenge is data integration (bringing together sequencing data + epi data), that will likely be easier from the epi side. If your primary goal is serving as a training and consultation resource to the epidemiologists, that could be addressed by a team sitting within the lab or epi. As the team grows these goals may change, but outlining the initial goals is an important step.
What are the roles of the genomic epidemiology team and how do they differ from the bioinformaticians and the existing epidemiologists
This one is key to both building a successful program, and strengthening the relationship between bioinformatics and epidemiology. When it is done right, bringing in a genomic epidemiologist should make the bioinformaticians jobs easier. When I was in a state-level genomic epi program, we were able to take on things like handling media inquiries, designing templates for metadata submission, designing a sentinel lab network and executing contracts with these labs for specimen submission, etc. These ‘program’ aspects may be something that your existing bioinformatics team enjoys, but often they are not high on the list. Again, this will be different based on your team and structure, so ask! If you have an existing bioinformatics team, ask what would functions and skills would help them do their job better and focus on their unique areas of expertise.
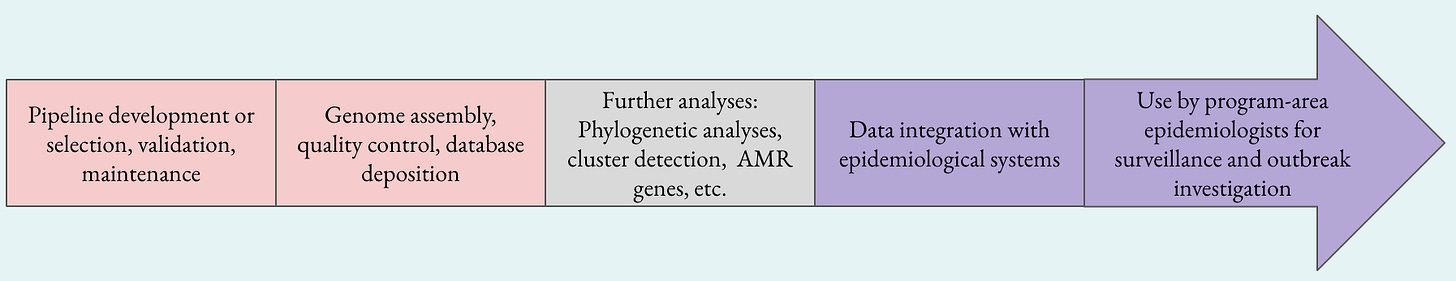
One clear distinction that should be made, is where the handover of data and responsibilities occur. This will be highly variable depending on the capabilities of your staff. In the diagram above on the left in pink are things that are generally clearly in the domain of the bioinformaticians, on the right are things that are in the domain of the epidemiologists. The gray box in the middle is doing a whole lot of work and could really be multiple boxes, but this is one of the key areas that needs to be explored and defined for each agency. Both bioinformatics and genomic epidemiology are highly interdisciplinary, and individual practitioners will have different skillsets and focuses. Just as bioinformaticians can vary in their balance of skillsets of computer science and biology, genomic epidemiologists vary in their skillset of applied epidemiology and bioinformatics. Titles don’t always fully define and convey individual background and skillsets!
Regardless of how the gray box activities are divided up, it is critical that those activities are closely tied in with the program-level epidemiologists who are the end users of the data. If the analyses don’t match the questions that the epidemiologists are trying to answer, the data will have much less impact. Working together with the end users to identify what is important can help guide both what the analyses should be, and who should do them.
Final thoughts
Regardless of how you structure your program, one key attribute that is required of your combined bioinformatics/genomic epidemiology workforce is a teaching spirit. There is so much to learn in these emerging fields, and generally there is never one person that has the training and experience to cover the full spectrum of activities that need to happen to make pathogen genomics a routinely integrated part of epidemiological surveillance and investigation in public health. Coming from a stronger field epidemiology background, I know that I need to lean on bioinformaticians for their expertise and learn from them. Two-way education needs to happen between your bioinformaticians and genomic epidemiologists, as well as from those groups out to program area epidemiologists, and local-level epidemiologists.
Thanks for reading! This post comes from my perspective and experience - I would love to hear from others. Interact with my posts on my linkedin page if you’ve like to join the conversation. If you’re looking to hire your first genomic epidemiologist and aren’t sure where to start, you can check out the table at www.genomicepi.com/jobs and download pdf files of job postings to get inspiration for a job description.


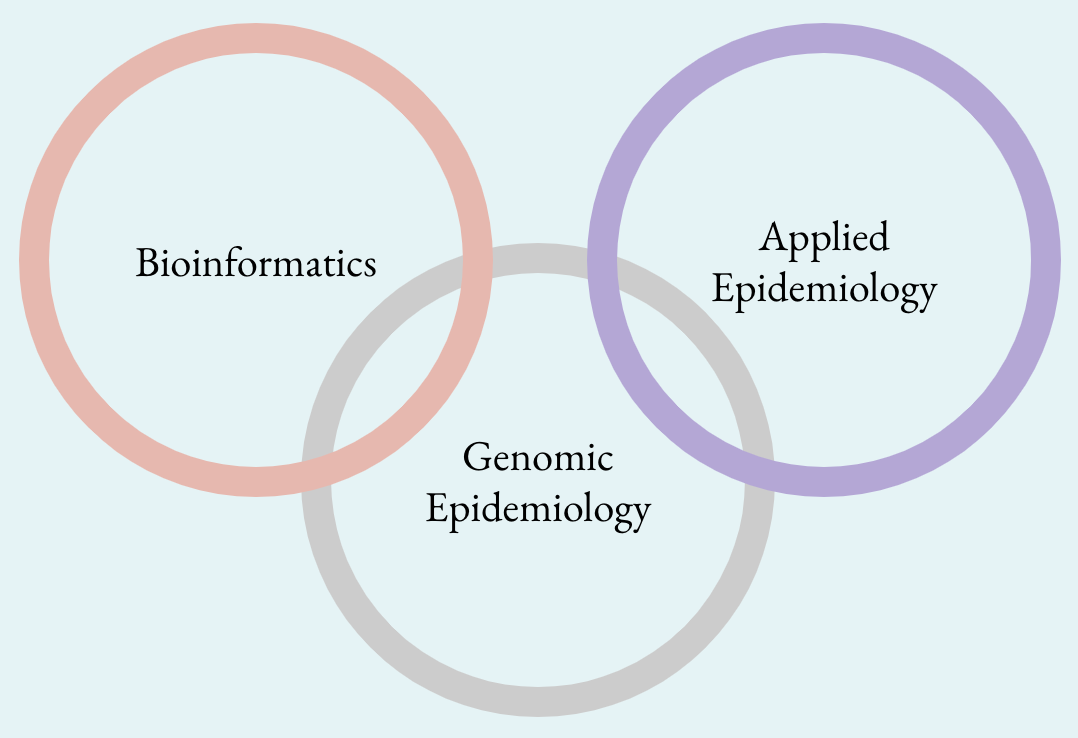
Very interesting view and questions about these "new jobs". I am microbiologist with expertise in bacterial genomics and I am doing the grey box in the middle! Most of the people are lost because I am not epidemiologist and I have only few skills in Bioinformatic BUT I can interpret the data and validate them as a microbiologist. Because I started my career (more than 30 years ago), I have a background on phenotypical traits that are expressed by bacteria. The good way of analyzing the data is to make the link between genotype and phenotype. In addition, I have a good knowledge in genetic of population specific of the different species that I am working on. This is not statistics (that is mainly used by epidemiologist) and this is not Bioinformatics (it's not about a script). In the middle, you have people that have experience in bacteria, the way of detecting, cultivate them, the way of storing, the way of testing them and the way of typing them...but I agree, things are going to change, I am almost a dinosaur now :-)